Track 1: Advances in Discovery, Formulation, and Delivery of New Modalities
Category: Poster Abstract
(T1030-02-06) A Mechanistic Model of IgG4-RD Identifies High-Potential Therapy Targets
- KS
Kalyanasundaram Subramanian
Differentia Bio
South San Francisco, California, United States - MG
Milad Ghomlaghi
AliveX Biotech
Grand Cayman, Cayman Islands - WZ
Wanbing Zhao
AliveX Biotech
Grand Cayman, Cayman Islands - HG
Hui Gao, Dr.
Peking University International Hospital
Beijing, Beijing, China (People's Republic) - CP
Cuiping Pan
Greater Bay Area Institute of Precision Medicine
Guangzhou, Guangdong, China (People's Republic) - AB
Aidin Biibosunov
Differentia Bio
South San Francisco, California, United States - YG
Yan Ge
AliveX Biotech
Grand Cayman, Cayman Islands - RS
Ruimin Sun
AliveX Biotech
Grand Cayman, Cayman Islands - DS
Di Sun
AliveX Biotech
Grand Cayman, Cayman Islands - XH
Xiaoqiang Hou
AliveX Biotech
Grand Cayman, Cayman Islands - WY
Wei Ye
AliveX Biotech
Grand Cayman, Cayman Islands - SW
Shirley Wen
AliveX Biotech
South San Francisco, California, United States - AK
Atefeh Kazeroonian, Ph.D. (she/her/hers)
Differentia Bio
South San Francisco, California, United States
Presenting Author(s)
Main Author(s)
Co-Author(s)
Purpose: IgG4-related disease (IgG4-RD) is a rare and newly discovered chronic inflammatory disease regulated by the immune system that affects 1 in 100,000 people per year. It is believed that immune disorders, genetics, infection, hypersensitivity and environmental factors are related to disease occurrence. The hallmarks of IgG4-RD include changes in levels of serum IgG4 and IgE, plasma cells and follicular helper T (Tfh) cells. Currently, the mechanisms underlying the occurrence and progression of IgG4-RD are not fully understood. This lack of mechanistic understanding has hindered development of biomarkers for early diagnosis as well as specific treatments for IgG4-RD[1,2,3]. In this study, we have developed an in silico model for IgG4-RD to provide a mechanistic understanding of disease pathology and to guide identification of therapy targets.
Methods: We collected PBMC samples from eight IgG4-RD patients and generated single-cell RNA sequencing, flow cytometry and cytokine profiling data. We first investigated the transcriptomic landscape of patient samples in comparison to healthy controls, and identified the cell types, proteins and inter-cellular communication patterns differentially regulated in IgG4-RD. We then employed our in-house knowledge graph to extract all the interactions between these cell types and cytokines, and developed an inter-cellular model topology for IgG4-RD. Using CERENA[4] and AMICI[5] packages, we developed an ordinary differential equations (ODE) model based on 1st and 2nd-order moment equations[6]. A multi-start gradient-based optimization algorithm implemented in PESTO[7] was employed to estimate the model parameters using cytokine profiling, flow cytometry and scRNA-seq data from patients. Finally, we used this model to test therapeutic strategies for IgG4-RD, by in silico perturbation of potential targets in the model.
Results: Through transcriptomics analysis, we captured changes in the immune system composition between IgG4-RD patient and healthy controls: we found enrichment of plasma B cell, Tregs and mDC populations, as well as reduction of naïve CD4 T cell population in IgG4-RD patients (Figure 1). In addition, patients showed enriched cell-cell communication in terms of overall interaction strength as well as specific patterns such as enhanced IL-10, IL-2, IL-4 and TGF-β signaling (Figure 2). Based on these findings, we developed an inter-cellular disease model that, among other immune cells, captures the interplay between Tfh and activated B cells, a dominant mechanism in IgG4-RD suggested by current literature. Our disease model comprises of more than thirty immune cell types and cytokines and around 100 kinetic parameters. The interactions between model components mirror both direct cell contact, as well as cytokine-mediated, activation and inhibition. Our model accurately reproduces the cell type percentages as well as the cytokine concentration data of patients (Figure 3). The estimated parameter values imply that although Tfh plays an important role in IgG4-RD, its rate of differentiation from activated CD4 T cells is much lower than other CD4 T subsets. Our results also suggest that IFN1, IL-21 and IL-6 have similar contributions to the differentiation of Tfh from CD4 T cells, while IL-4 is the dominant activating factor for B cells. In silico perturbation experiments based on our model suggest that targeting IL-4 could partially reverse the disease hallmarks, such as the elevated level of Tfh and activated CD4 T cells. Inspecting our single-cell RNAseq data, we found enriched interactions mediated by IL-4 in the patient population, supporting this model prediction at the molecular level.
Conclusion: Our calibrated disease model hints at mechanistic differences between patients and healthy controls, and sheds light on cellular dynamics that are dysregulated in IgG4-RD. This model can also be used to propose novel therapeutic targets for IgG4-RD by simulating the effects of potential target perturbations on the disease endpoint, such as the IgG4 serum level. Although wet-lab and clinical validations remain necessary to confirm targets identified by the model, we were able to find supporting evidence in our transcriptomic data. This model can further utilize PD data in clinical trials to form a QSP model that establishes an accurate relationship between drug exposure and clinical outcome informed by the disease biology. In this way, our model can enhance success rates in the later stages of drug development for IgG4-RD.
References: [1] Karadeniz H, Vaglio A. IgG4-related disease: a contemporary review. Turk J Med Sci. 2020 Nov 3;50(SI-2):1616-1631. doi: 10.3906/sag-2006-375. PMID: 32777900; PMCID: PMC7672352.
[2] Opriţă R, Opriţă B, Berceanu D, Diaconescu IB. Overview of IgG4 - Related Disease. J Med Life. 2017 Oct-Dec;10(4):203-207. PMID: 29362594; PMCID: PMC5771249.
[3] Wu X, Peng Y, Li J, Zhang P, Liu Z, Lu H, Peng L, Zhou J, Fei Y, Zeng X, Zhao Y, Zhang W. Single-Cell Sequencing of Immune Cell Heterogeneity in IgG4-Related Disease. Front Immunol. 2022 May 27;13:904288. doi: 10.3389/fimmu.2022.904288. PMID: 35693817; PMCID: PMC9184520.
[4] A. Kazeroonian, F. Fröhlich, A. Raue, F. J. Theis, J. Hasenauer. CERENA: ChEm- ical REaction Network Analyzer – A Toolbox for the Simulation and Anal- ysis of Stochastic Chemical Kinetics. PLOS ONE 11(1): e0146732, 2016.
[5] Fröhlich, F., Weindl, D., Schälte, Y., Pathirana, D., Paszkowski, Ł., Lines, G.T., Stapor, P. and Hasenauer, J., 2021. AMICI: High-Performance Sensitivity Analysis for Large Ordinary Differential Equation Models. Bioinformatics, btab227, DOI:10.1093/bioinformatics/btab227.
[6] S. Engblom. Computing the moments of high dimensional solutions of the master equation. Appl. Math. Comp., 180:498–515, 2006. doi: 10.1016/j.amc.2005.12.032.
[7] Stapor, P., Weindl, D., Ballnus, B., Hug, S., Loos, C., Fiedler, A., Krause, S., Hross, S., Fröhlich, F., Hasenauer, J. (2018). PESTO: Parameter EStimation TOolbox. Bioinformatics, 34(4), 705-707. doi: 10.1093/bioinformatics/btx676.
.jpg) Figure 1: Transcriptomic analysis of patient and control single-cell RNA sequencing data reveals the percentage of various immune cells in each group. Each column represents a subject in the corresponding group
Figure 1: Transcriptomic analysis of patient and control single-cell RNA sequencing data reveals the percentage of various immune cells in each group. Each column represents a subject in the corresponding group.jpg) Figure 2: The information flow due to various signalling networks in patient and control groups. The incoming and outgoing signalling mediated by various cytokines were analysed based on scRNAseq data, and the corresponding information flow strength was compared between patients (blue) and healthy controls (red). The quantification of information flow and cell-cell communication was done in the CellChat package.
Figure 2: The information flow due to various signalling networks in patient and control groups. The incoming and outgoing signalling mediated by various cytokines were analysed based on scRNAseq data, and the corresponding information flow strength was compared between patients (blue) and healthy controls (red). The quantification of information flow and cell-cell communication was done in the CellChat package.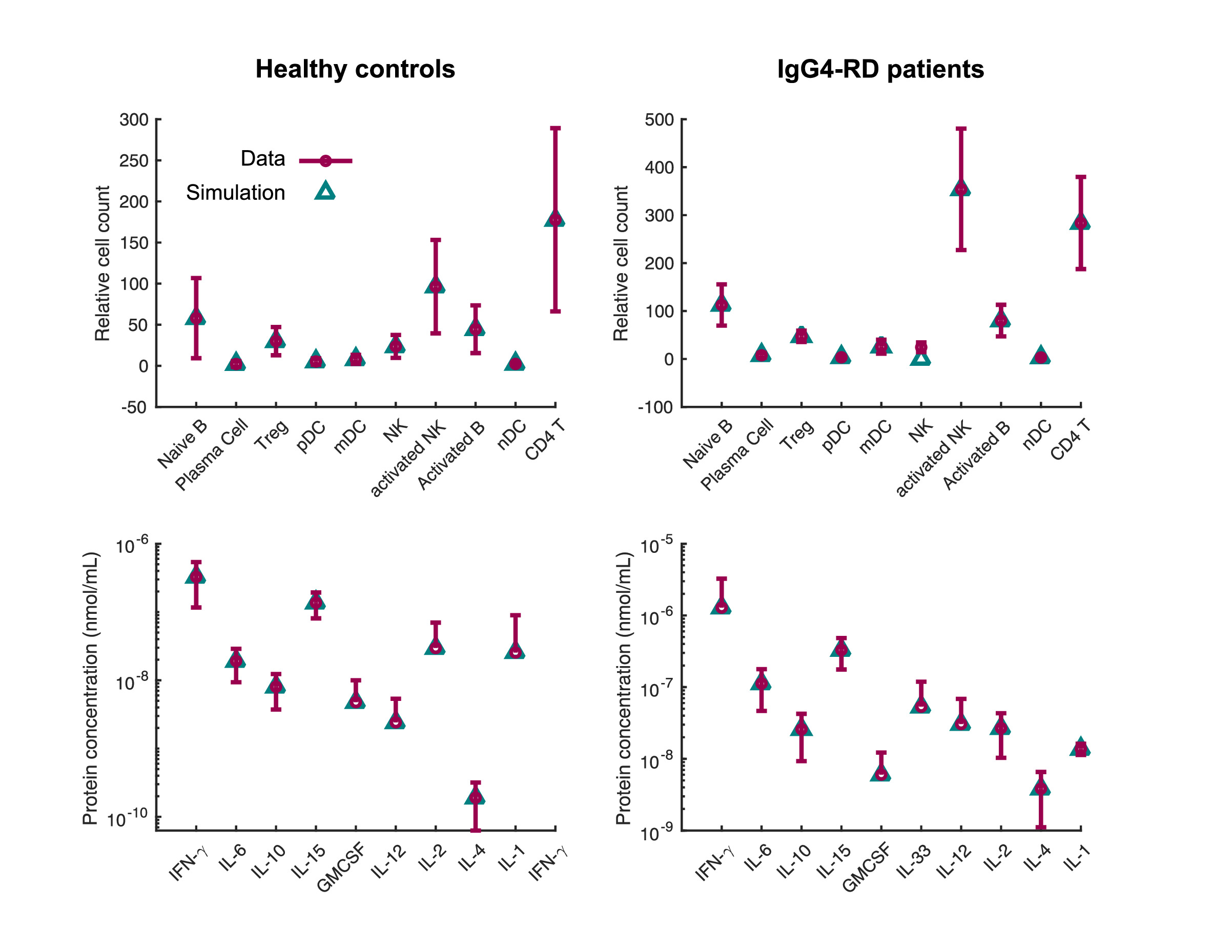 Figure 3: The calibrated IgG4-RD disease model reproduces the steady state data of patient and control groups. The comparison of model simulations against experimental data is shown for relative cell counts (top row) and protein concentrations (bottom row). The parameters of the disease model were (partly) estimated to fit the data.
Figure 3: The calibrated IgG4-RD disease model reproduces the steady state data of patient and control groups. The comparison of model simulations against experimental data is shown for relative cell counts (top row) and protein concentrations (bottom row). The parameters of the disease model were (partly) estimated to fit the data.